The GREGoR Dataset includes a wide range of data and file formats as described in the GREGoR Data Model. Analysts working with GREGoR Data can find file paths to molecular data files using the structure of the Data Model and search features within an AnVIL workspace to find specific information in workspace data tables. We have also compiled the following information about how to find certain files of broad interest. Note: if you have not already done so, please first follow the instructions at the Getting Started section of the Getting Started on AnVIL with GREGoR webpage to set-up your AnVIL account and secure data access permissions.
Locations of key files
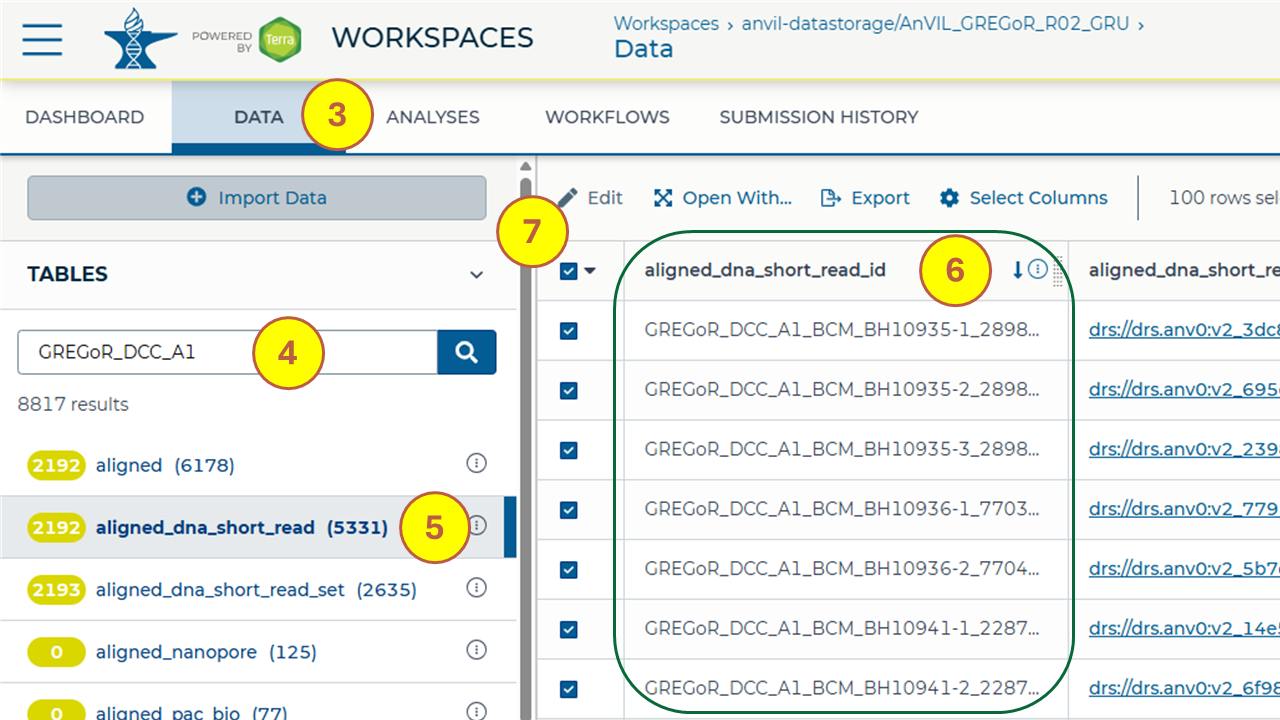
Refer to the table below for the information needed to find key files of interest in an AnVIL workspace (step numbers are annotated in the screenshot and specified in the table below).
- Log in to AnVIL at anvil.terra.bio.
- Select “Workspaces” from the menu in the upper left. Navigate to a workspace of interest (recommended to choose the latest released GREGoR workspace, e.g. AnVIL_GREGoR_R**_GRU or AnVIL_GREGoR_R**_HMB).
- Navigate to the “DATA” tab from the menu bar at the top.
- Search for the text string - as specified in the table below - by entering into the search box under the “TABLES” section in the left sidebar to search across all data tables in the workspace.
- Filter your results by selecting the data table - as specified in the table below - from the left sidebar (which you may need to expand).
- Refer to the column - as specified in the table below - and identify rows with the matching text string. You might need to expand the columns in the data table individually to find the specific column of interest. Rows that contain the matching text string in the specified column include paths to the key files of interest.
- Select the checkboxes for all the files that match the search criteria to open or export.
Examples of querying the AnVIL workspace for these files using R are available in this R notebook. For resources on interacting with and analyzing GREGoR data, refer to the AnVIL Resources webpage.
| Key file(s) | Description of the file(s) | Step 4: Enter text string into search box | Step 5: Select data table | Steps 6 & 7: Refer to column & select (check) rows including text string |
|---|---|---|---|---|
| Harmonized CRAMs | Alignment files reprocessed by the GREGoR DCC | GREGoR_DCC_A1 | aligned_dna_short_read | aligned_dna_short_read_id = “GREGoR_DCC_A1*” (select all rows) |
| GVCFs | Single sample genomic VCFs generated by the DCC from harmonized CRAMs | gVCF | called_variants_dna_short_read | analysis_details = “gVCF generated with*“ (select all rows) |
| GREGoR joint callset: Unannotated & Annotated VCFs | Unannotated and VEP annotated multi-sample chromosome VCFs generated by the DCC from single sample GVCFs | consortium-wide callset | called_variants_dna_short_read | Annotated VCF: Unannotated VCF: |
| Broad de novo callset: Complete & Confident set | Complete and confident sets of a Consortium-wide de novo callset provided by the Broad | GREGoR_DENOVO | called_variants_dna_short_read | Confident set: aligned_dna_short_read_set_id = “GREGoR_DENOVO_CONFIDENT*” Complete set: aligned_dna_short_read_set_id = “GREGoR_DENOVO_S*” |
| Broad mitochondrial callset | Consortium-wide mitochondrial callset provided by the Broad | Broad_U07_GS_mtDNA_callset | called_variants_dna_short_read | ali gned_dna_short_read_set_id = “Broad_U07_GS_mtDNA_callset*" |
