Date Approved: January 18, 2023
Date of Last Update: January 15, 2025
Version: 1.4
Background
The central mission of the GREGoR (Genomics Research to Elucidate the Genetics of Rare Diseases) Consortium is to significantly increase the proportion of Mendelian conditions with an identified genetic cause. To advance this mission, GREGoR aims to create datasets with broad utility and disseminate beyond the consortium through data sharing. The GREGoR consortium is committed to comprehensive and rapid data sharing of “GREGoR Data” (or “Data”) which includes the following data types: phenotypic, genotyping, nucleic acid sequence, multi-omic, biomarker, and any other "Scientific Data" as defined in the Final NIH Policy for Data Management and Sharing (see section 'Definition of “Scientific Data”'). It also includes preliminary analyses, re-processed, derived, and associated data generated from samples collected and/or sequenced under the primary GREGoR grant mechanisms across the consortium. For the purposes of this policy, “external” refers to sites and resources used by the broader scientific community and not just the GREGoR consortium.
Back to topScope
The External Data Sharing Policy is designed to describe the required and recommended external data sharing strategies to be followed by Research Centers (RCs) within the Consortium, and by extension Research Center Collaborators who make use of GREGoR resources to generate data or results. This policy establishes and documents minimum data sharing requirements for the Consortium and supports the scope of GREGoR RC’s Institutional Review Board (IRB) protocols, GREGoR members, GREGoR partner members, and GREGoR collaborators who contribute to the generation or analyses of these data are expected to follow the GREGoR External Data Sharing Policy.
Back to topPolicy
Minimum Standards and Timeline
All required external data sharing sites, including GREGoR’s minimum standards, timelines and groups responsible for deposition, are listed in Table 1. The GREGoR Consortium recognizes that non-GREGoR resources (listed in table below) will vary in their minimum requirements to share data; however, GREGoR standards for sharing will frequently be higher. The Consortium will meet the resource-specified minimum requirements listed below, but also strive to include as much additional data as our data model allows.
Table 1. Required External Data Sharing Sites
| Resource | GREGoR’s Minimum Expectation for Data Shared | Timeline for Sharing | Group responsible for deposition |
| GREGoR Website | Cohort level descriptions, overview of data sharing activities with links on how to find that data in sources listed below | Cohorts: as cases/cohorts enter the study, no less than quarterly | Data Coordinating Center with information provided by Research Centers |
| AnVIL (non-internal workspaces) | All required fields in data model, including data files | 1 month after RC’s data submission the DCC will inform AnVIL that the workspace is approved for release | Data Coordinating Center with data provided by Research Centers |
| Matchmaker Exchange (MME) nodes | Gene, Variants, Phenotype | Ideally as candidates are identified, and no less than quarterly | Research Centers |
| ClinVar | Variant, gene, disease (ideally Mondo term), classification and evidence (including case-level phenotype, ACMG/AMP evidence codes, PubMed IDs)* |
Variants being included with publication should be submitted before/with manuscript
|
Research Centers |
| GenCC | Novel or Corrected Gene-Disease relationships |
Gene-disease relationships being included with publication should be submitted or updated before/with manuscript For gene-disease relationships not scheduled for publication, submit to GenCC at least quarterly |
Research Centers |
*Containing enough information to follow how you arrived at your classification (for example, this could include a descriptive summary, and/or submitting ACMG/AMP criteria codes and PMIDs)
Prioritization of ClinVar Variant Submissions
Table 2. Prioritization of ClinVar Variant Submissions
| Variant Type | Priority | Prioritization Notes |
|
Known disease genes |
High
Low |
Definitive/Strong/Moderate in GenCC, or likely to reach that classification if not already curated:
|
|
Novel disease gene relationships |
High |
Limited or below in GenCC, or likely to reach that classification if not already curated:
|
Tracking/Reporting
To minimize manual reporting of data sharing activities, when possible, tracking of submissions to each of the required sites listed in Table 1 will be performed via the following metrics:
- AnVIL (non-internal workspaces): Tracking of which workspaces have been made public will be maintained by the DCC and shared with NHGRI on a regular basis.
- MME: RCs will report their number of submissions to Matchmaker Exchange (via their node of choice) to the DCC as part of the regular data reporting processes for each upload cycle.
- ClinVar: RCs must add “GREGoR Consortium” to the study name in the ClinVar submission portal (see image below). Additionally, RCs must provide ClinVar SCV IDs when formatting their dataset(s) to the GREGoR Data model during uploads for Consortium data sharing, which will link to the latest classification.
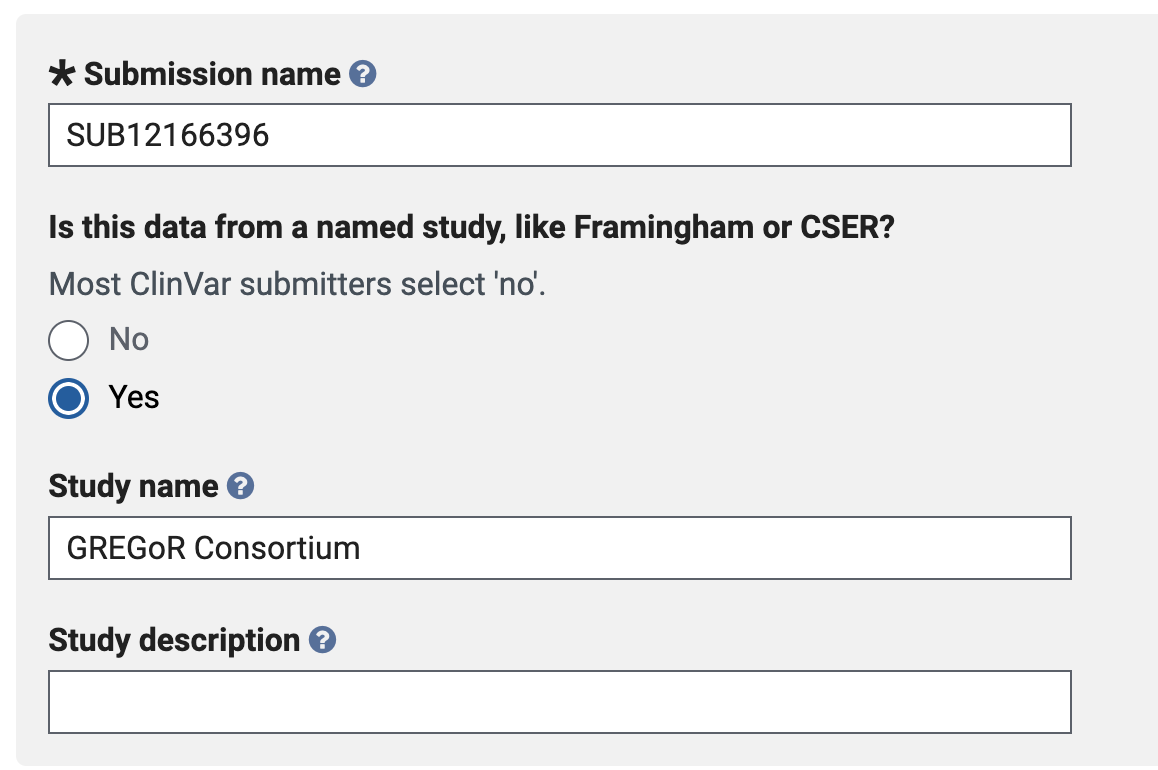
- GenCC: RC will report their number of submissions to GenCC (via their profile of choice) to the DCC as part of regular data reporting processes for each upload cycle.
Future Directions
The Consortium recognizes that evolving needs for interoperability with ongoing and planned federated networks (e.g. Matchmaker Exchange) may influence the recommended data structure of GREGoR shared data. Additionally, any new data types required or requested by external sites, but not currently included in our data model, will be assessed by the Data Standards & Analysis Working Group to determine if the Consortium can reasonably include this data in future data models.
Back to topRelated Policies
- GREGoR Data Sharing Agreement
- Investigator Collaboration Agreement
- GREGoR AnVIL data model V1
- NIH Policy for Data Management and Sharing
Definitions
- “GREGoR Data” (or “Data”) - (As per our Consortium Data Sharing Agreement) Includes phenotypic data, genotyping data, nucleic acid sequence data, multi-omic, biomarker, "Scientific Data" as defined in the Final NIH Policy for Data Management and Sharing (see section 'Definition of “Scientific Data”'), preliminary analyses, re-processed, derived, and associated metadata generated from samples collected and/or sequenced under the primary GREGoR grant mechanisms outlined above and that is shared under this Agreement.
Change Log
- V1.4 - Revision to tracking/reporting requirements based on implemented processes for reporting data submissions to external repositories starting U08 and further refined in discussions with RCs 01/2025
- V1.3 - Revision based on Steering Committee discussion on 1/18/2023 regarding minimum data standards for external repositories
- V1.2 - Revision based on Policy WG discussion 8/24/2022, with additional edits/comments/notes from 9/14/2022 Policy WG discussion
- V1.1 - Includes edits/notes from Policy WG discussion 8/24/2022
- V1.0 - Initial Policy
