GREGoR Consortium Research Centers and Partners collect clinical, phenotype, and molecular data that is combined to form the GREGoR Dataset. The GREGoR Dataset is registered with the database of Genotypes and Phenotypes (dbGaP) and released to the scientific community via controlled access on NHGRI’s AnVIL platform. The dbGaP study webpage for GREGoR (phs003047) includes additional study design and enrollment information.
Back to topThe GREGoR Dataset
The GREGoR Dataset includes family structure, phenotype, case status, genetics findings, short read whole exome sequencing (WES), whole genome sequencing (WGS), long read WGS, and short read RNA-seq data. It also includes uniformly processed short-read WGS files and a joint callset of single nucleotide variants and short insertions and deletions. Future data releases will include additional molecular data types and information.
GREGoR Dataset characteristics by release version
| GREGoR release version | R01 | R02 | R03 | R04 |
|---|---|---|---|---|
| dbGap accession | phs003047.v1 | phs003047.v2 | phs003047.v3 | phs003047.v4 |
| Release date | September 2023 | November 2024 | July 2025 | October 2025 |
| Number of participants Number of families Consent groups1 | 2,512 990 GRU, HMB | 7,394 3,059 GRU, HMB | 8,840 3,610 GRU, HMB | 10,683 4,366 GRU, HMB |
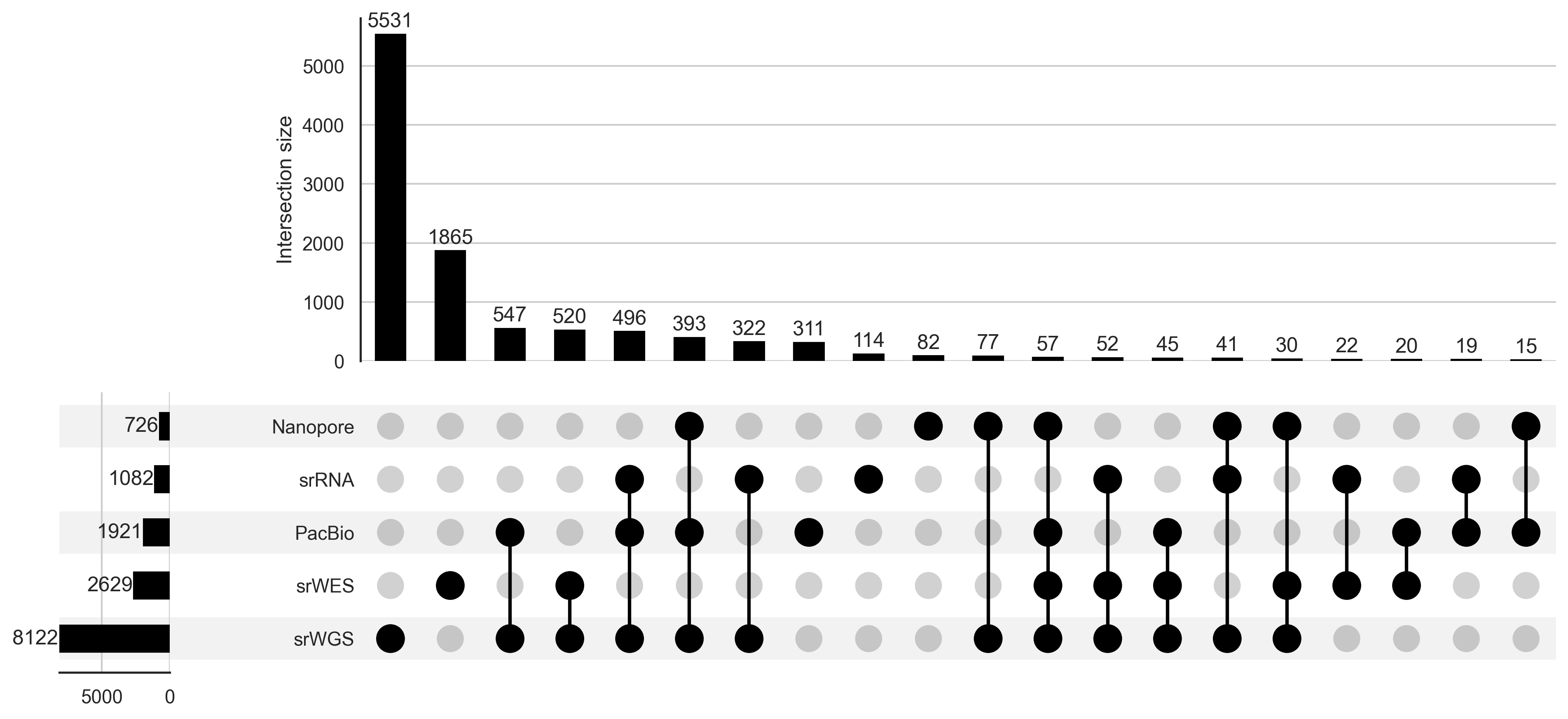
| Available experimental data2 Short read WGS Short read WES Long read WGS Short read RNA-seq Short read ATAC-seq | 1,441 997 0 183 0 | 5,182 2,242 214 539 0 | 6,535 2,284 1,772 860 189 | 8,161 2,629 2,648 1,100 189 |
| Size (TB) | 61.8 | 149.5 | 350.3 | 546.5 |
| Joint callset (SNVs, short indels)3 Number of samples Size4 Sites-only files | 2,353 ~84 GiB | 2,351 ~84 GiB R03 folder | 3,624 ~129 GiB R04 folder | |
| GREGoR Data Model version | 1.1 | 1.6 | 1.7 | 1.9 |
| Genome build | GRCh38 | GRCh38 | GRCh38 | GRCh38 |
| Methods documentation | R01 pdf | R02 pdf | R03 pdf | R04 pdf |
| Additional dataset characteristics | R02 pdf | R03 pdf | R04 pdf | |
| Release Notes | R01 pdf | R02 pdf | R03 pdf | R04 pdf |
| Errata | R02 | R03 | R04 |
1GRU: General Research Use; HMB: Health/Medical/Biomedical
2WGS: Whole Genome Sequencing; WES: Whole Exome Sequencing
3SNVs, indels: Single Nucleotide Variants and short insertions and deletions
4Joint called VCFs are separated by chromosome; size is for all chromosomes together
Overlapping -omics data in GREGoR
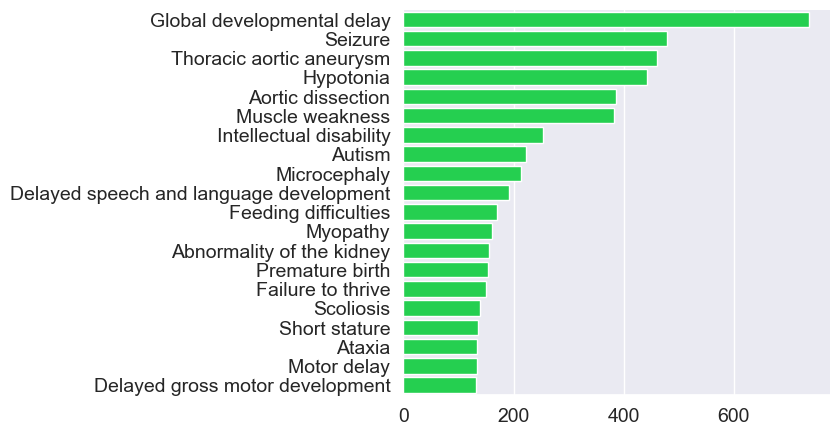
Phenotypes in the GREGoR Dataset
 Back to top
Back to top
Accessing GREGoR data
Researchers from the scientific community can apply for controlled access to GREGoR data stored on the AnVIL platform via dbGaP. Applicants should submit Data Access Requests (DARs) by selecting datasets associated with dbGaP accession number phs003047 by following the NIH Scientific Data Sharing instructions for How to Request and Access Datasets from dbGaP.
Alternatively, the GREGoR Consortium offers a Partner Membership opportunity to enable investigators to actively participate in and contribute to the scientific activities and mission of the GREGoR Consortium. For more information, please see the GREGoR Partner Members webpage.
Back to topWorking with GREGoR data
AnVIL is the primary repository for GREGoR data. AnVIL provides controlled-access data storage and a cloud-based analysis environment for researchers. Links to set-up accounts and billing on AnVIL, platform tutorials, how to find files, and related resources are available at Getting Started on AnVIL with GREGoR .
The GREGoR Consortium also regularly contributes data to several resources (e.g. Variant Browser, ClinVar, GenCC, MME) that support rare disease research which are described in Tools and Resources to interact with GREGoR data.
Back to topFuture Data Releases
The GREGoR Consortium continues to collect and aggregate participant, family and phenotype data, as well as a range of molecular data to support rare disease research and to share with the scientific community. GREGoR Research Centers and Partner Members are expanding the GREGoR Data Model to support additional data types, including metabolomics, and more.
The GREGoR Dataset will continue to expand with additional participant, family, phenotype and other data types available for analysis. We anticipate that periodic releases will continue throughout the life of the Consortium, and will remain in NIH data repositories as a valuable scientific resource.
Back to topAdditional resources
- GREGoR at ASHG
- GREGoR Publications
- GREGoR datasets on AnVIL Data Explorer
- GREGoR dbGaP study webpage - phs003047
- GREGoR DUOS study webpage - phs003047
Please provide feedback on the GREGoR Dataset!
We are very interested in making the GREGoR Dataset broadly useful. Please let us know how we’re doing or how we might improve the dataset by contacting the GREGoR Data Coordinating Center (select “Data” from the Topic dropdown menu).
Back to top